Homework 8
Maddie Winer
2025-03-19
Question 1
Copy and paste the code below into different code chunks, and then read the text and run the code chunks one at a time to see what they do.
#open libraries
library(ggplot2)
library(MASS) ##
## Attaching package: 'MASS'## The following object is masked from 'package:dplyr':
##
## select#read in data vector
z <- rnorm(n=3000,mean=0.2)
z <- data.frame(1:3000,z)
names(z) <- list("ID","myVar")
z <- z[z$myVar>0,]
str(z)## 'data.frame': 1750 obs. of 2 variables:
## $ ID : int 1 2 3 4 5 6 11 13 14 17 ...
## $ myVar: num 0.725 1.401 0.674 1.616 0.489 ...summary(z$myVar)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.000487 0.355624 0.760747 0.882274 1.264636 4.312080#plot histogram
p1 <- ggplot(data=z, aes(x=myVar, y=..density..)) +
geom_histogram(color="grey60",fill="cornsilk",linewidth=0.2)
print(p1)## Warning: The dot-dot notation (`..density..`) was deprecated in ggplot2
## 3.4.0.
## ℹ Please use `after_stat(density)` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this
## warning was generated.## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.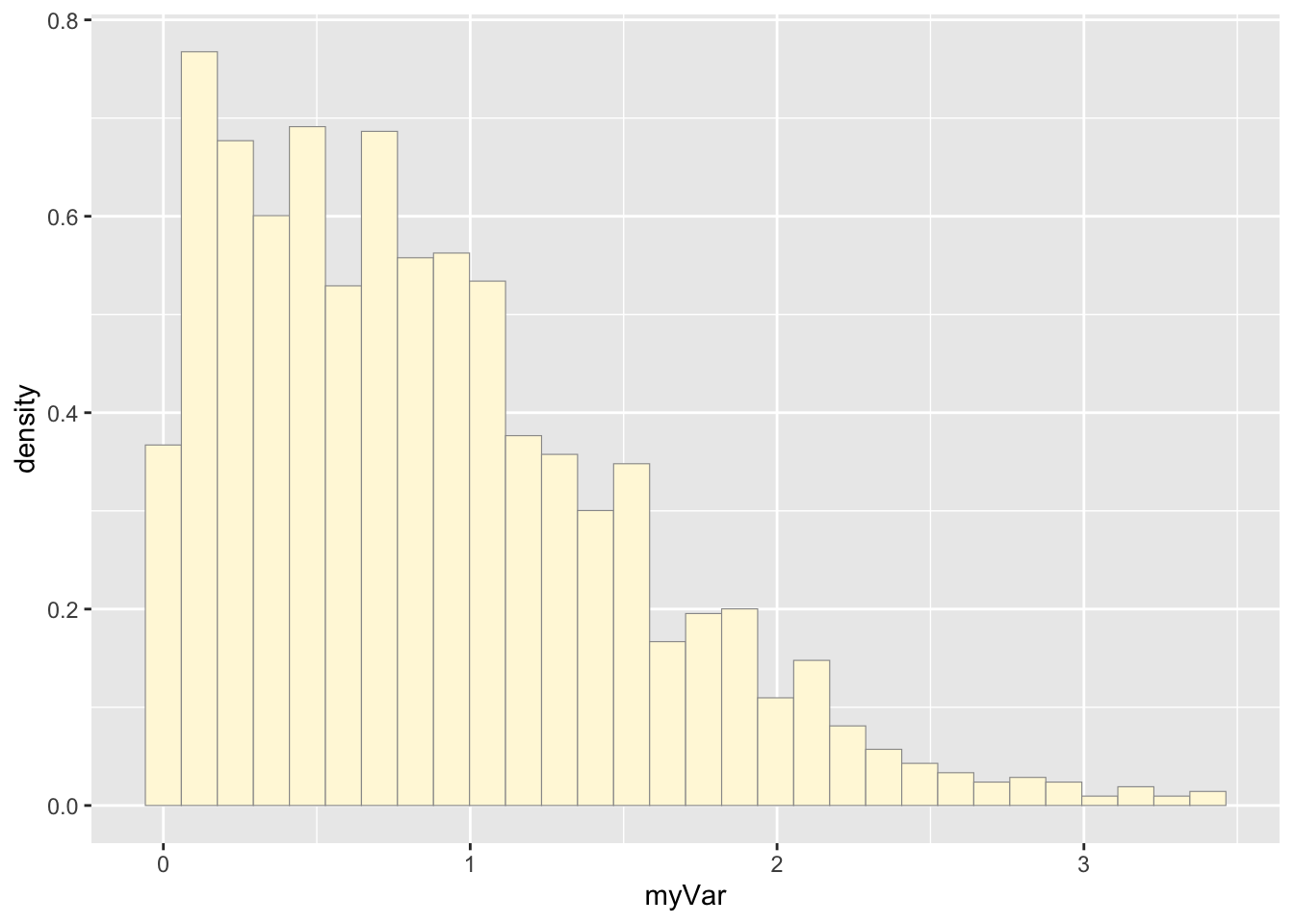
#add empirical density curve
p1 <- p1 + geom_density(linetype="dotted",size=0.75)## Warning: Using `size` aesthetic for lines was deprecated in ggplot2
## 3.4.0.
## ℹ Please use `linewidth` instead.
## This warning is displayed once every 8 hours.
## Call `lifecycle::last_lifecycle_warnings()` to see where this
## warning was generated.print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.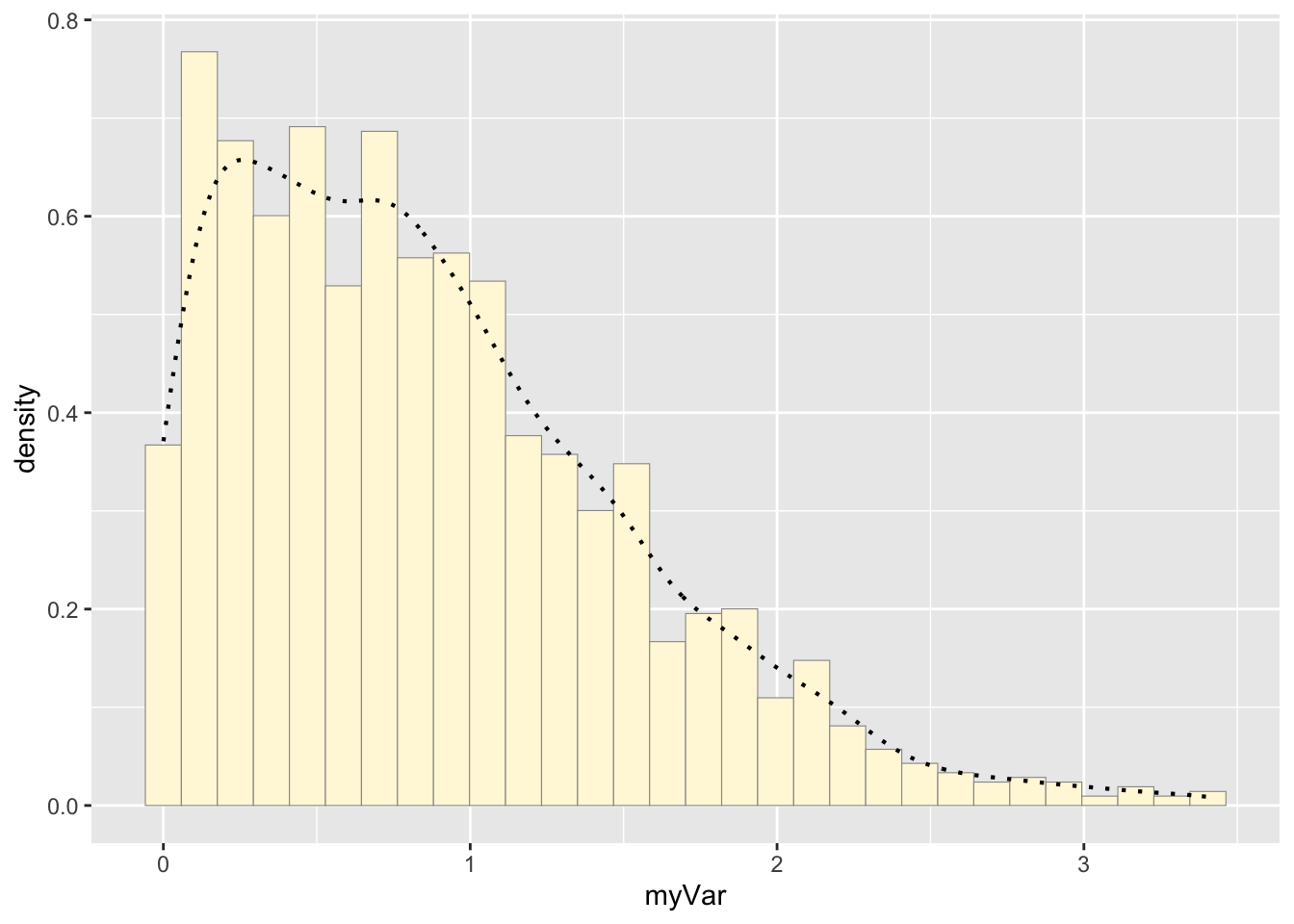
#get maximum likelihood parameters for normal
normPars <- fitdistr(z$myVar,"normal")
print(normPars)## mean sd
## 0.88227449 0.65839142
## (0.01573857) (0.01112885)str(normPars)## List of 5
## $ estimate: Named num [1:2] 0.882 0.658
## ..- attr(*, "names")= chr [1:2] "mean" "sd"
## $ sd : Named num [1:2] 0.0157 0.0111
## ..- attr(*, "names")= chr [1:2] "mean" "sd"
## $ vcov : num [1:2, 1:2] 0.000248 0 0 0.000124
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:2] "mean" "sd"
## .. ..$ : chr [1:2] "mean" "sd"
## $ n : int 1750
## $ loglik : num -1752
## - attr(*, "class")= chr "fitdistr"normPars$estimate["mean"] ## mean
## 0.8822745#plot normal probability density
meanML <- normPars$estimate["mean"]
sdML <- normPars$estimate["sd"]
xval <- seq(0,max(z$myVar),len=length(z$myVar))
stat <- stat_function(aes(x = xval, y = ..y..), fun = dnorm, colour="red", n = length(z$myVar), args = list(mean = meanML, sd = sdML))
p1 + stat## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.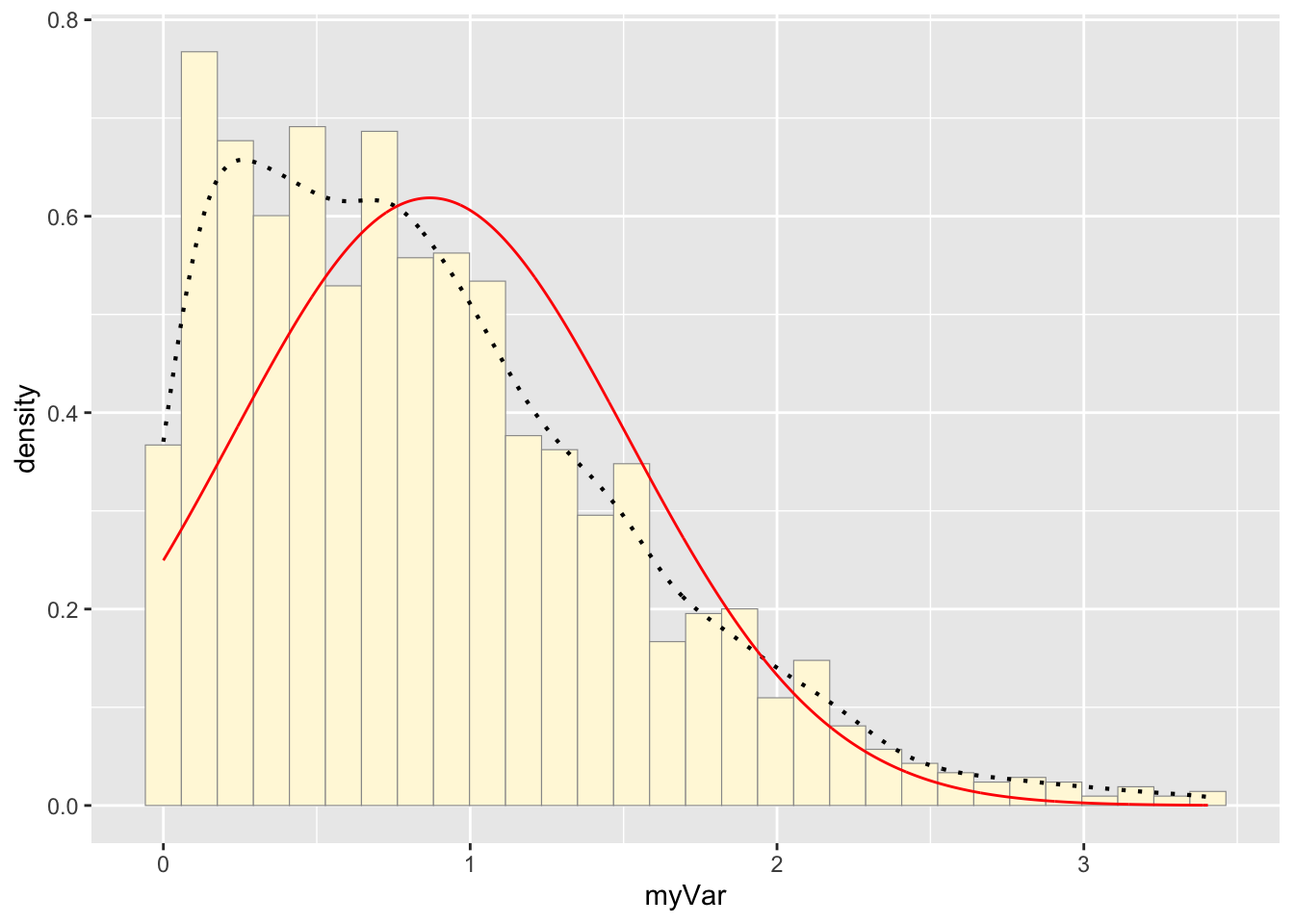
#plot exponential probability density
expoPars <- fitdistr(z$myVar,"exponential")
rateML <- expoPars$estimate["rate"]
stat2 <- stat_function(aes(x = xval, y = ..y..), fun = dexp, colour="blue", n = length(z$myVar), args = list(rate=rateML))
p1 + stat + stat2## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.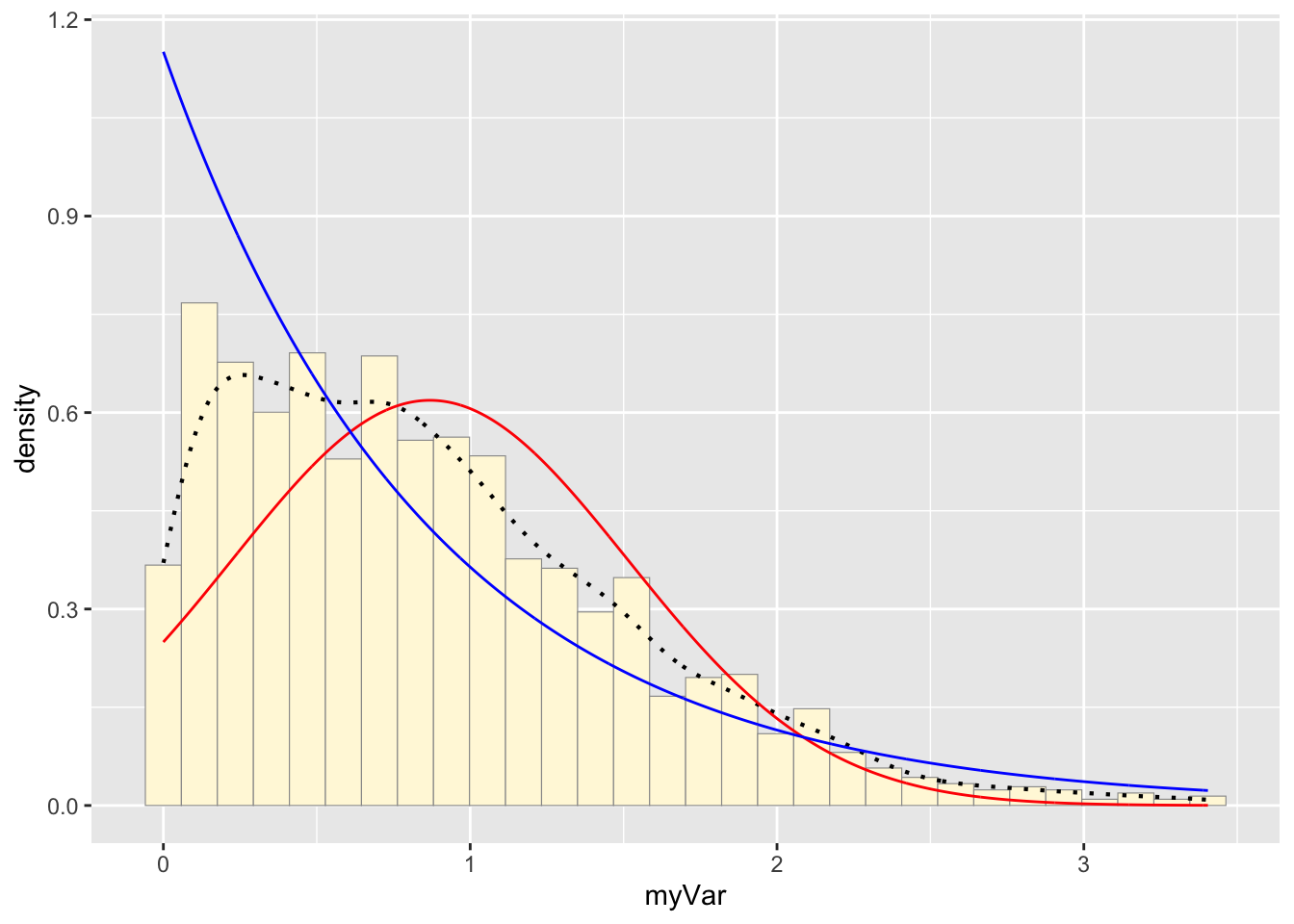
#plot uniform probability density
stat3 <- stat_function(aes(x = xval, y = ..y..), fun = dunif, colour="darkgreen", n = length(z$myVar), args = list(min=min(z$myVar), max=max(z$myVar)))
p1 + stat + stat2 + stat3## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.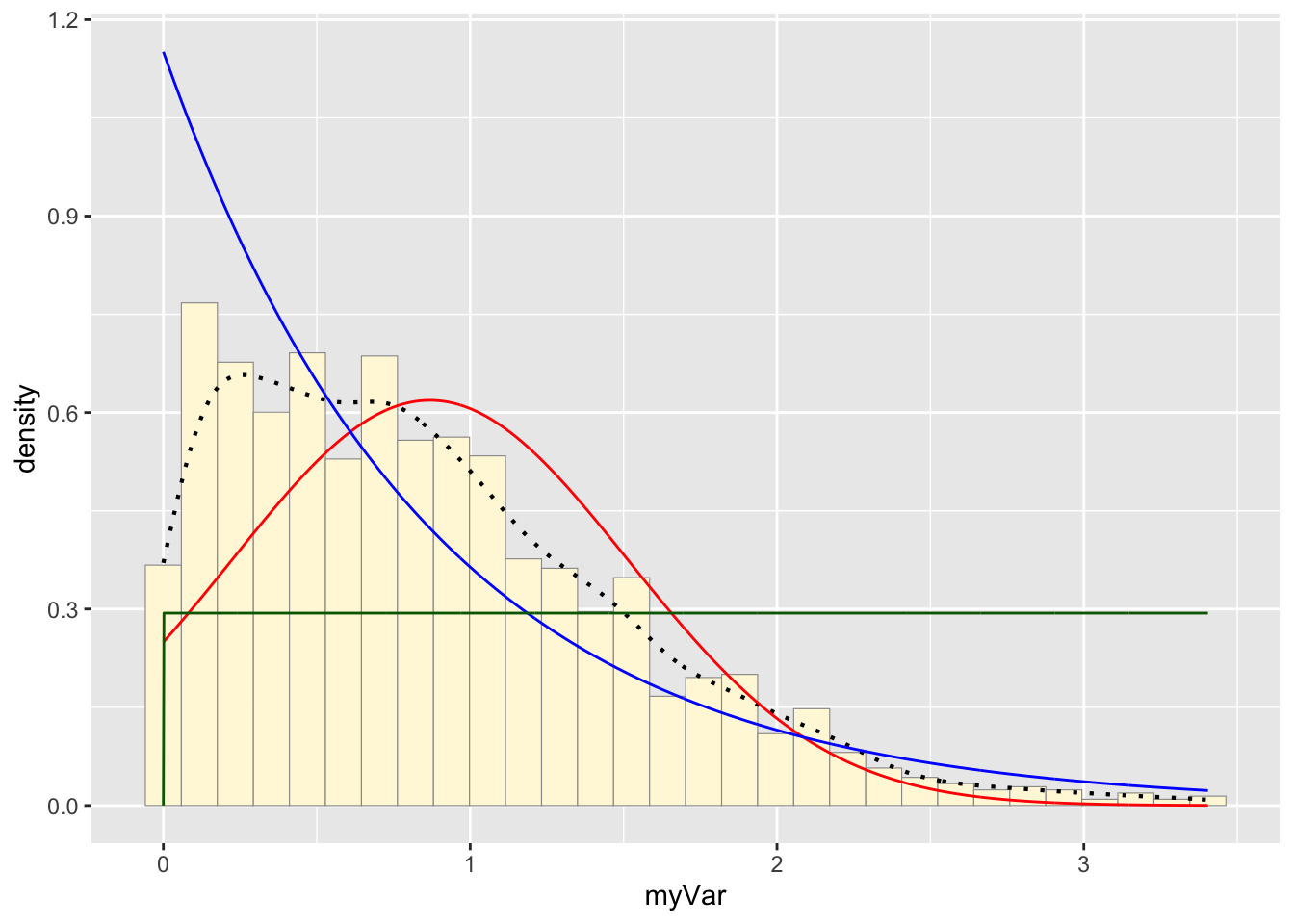
#plot gamma probability density
gammaPars <- fitdistr(z$myVar,"gamma")## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs producedshapeML <- gammaPars$estimate["shape"]
rateML <- gammaPars$estimate["rate"]
stat4 <- stat_function(aes(x = xval, y = ..y..), fun = dgamma, colour="brown", n = length(z$myVar), args = list(shape=shapeML, rate=rateML))
p1 + stat + stat2 + stat3 + stat4## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.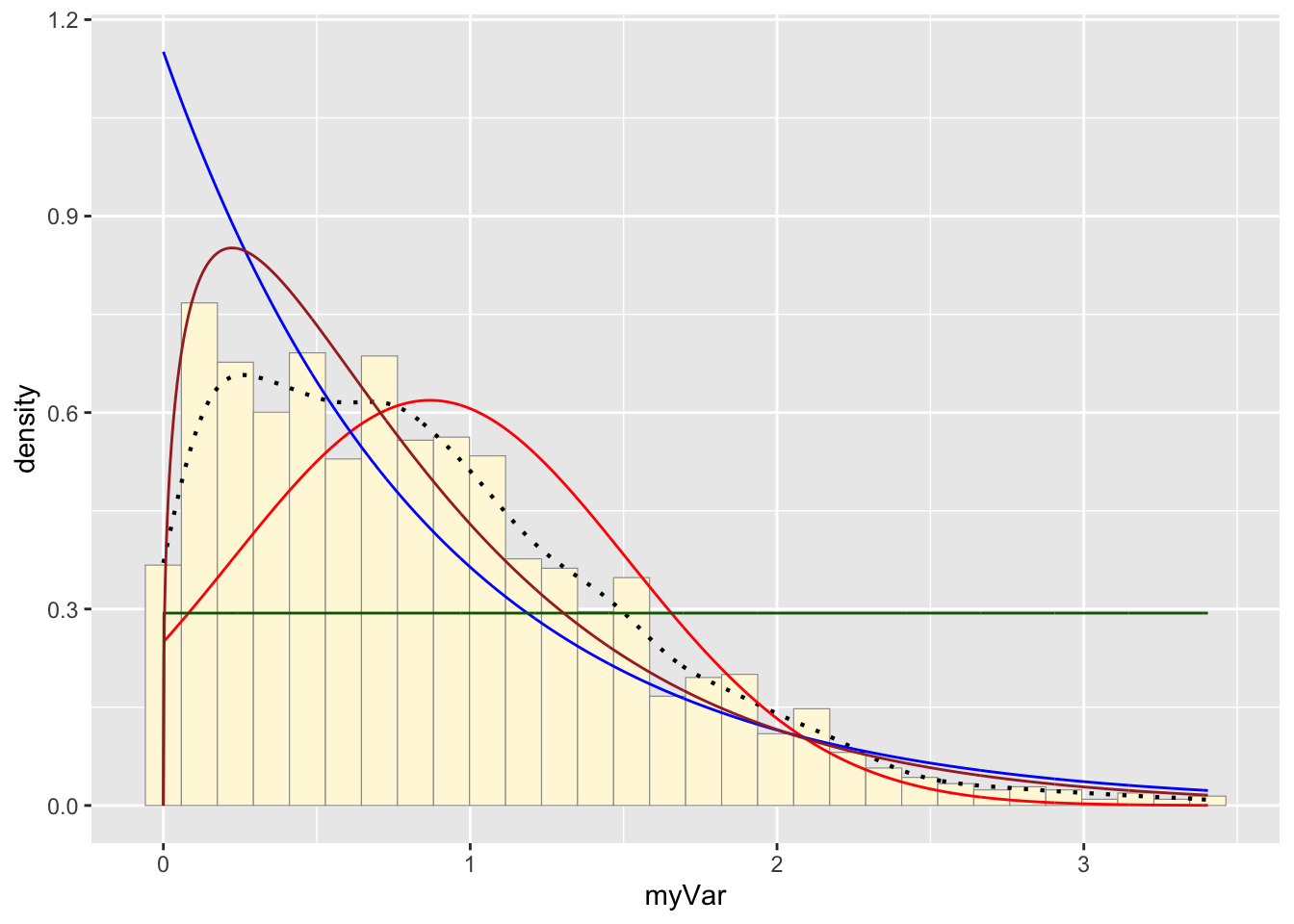
#plot beta probability density
pSpecial <- ggplot(data=z, aes(x=myVar/(max(myVar + 0.1)), y=..density..)) +
geom_histogram(color="grey60",fill="cornsilk",size=0.2) +
xlim(c(0,1)) +
geom_density(size=0.75,linetype="dotted")
betaPars <- fitdistr(x=z$myVar/max(z$myVar + 0.1),start=list(shape1=1,shape2=2),"beta")## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs producedshape1ML <- betaPars$estimate["shape1"]
shape2ML <- betaPars$estimate["shape2"]
statSpecial <- stat_function(aes(x = xval, y = ..y..), fun = dbeta, colour="orchid", n = length(z$myVar), args = list(shape1=shape1ML,shape2=shape2ML))
pSpecial + statSpecial## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.## Warning: Removed 2 rows containing missing values or values outside the
## scale range (`geom_bar()`).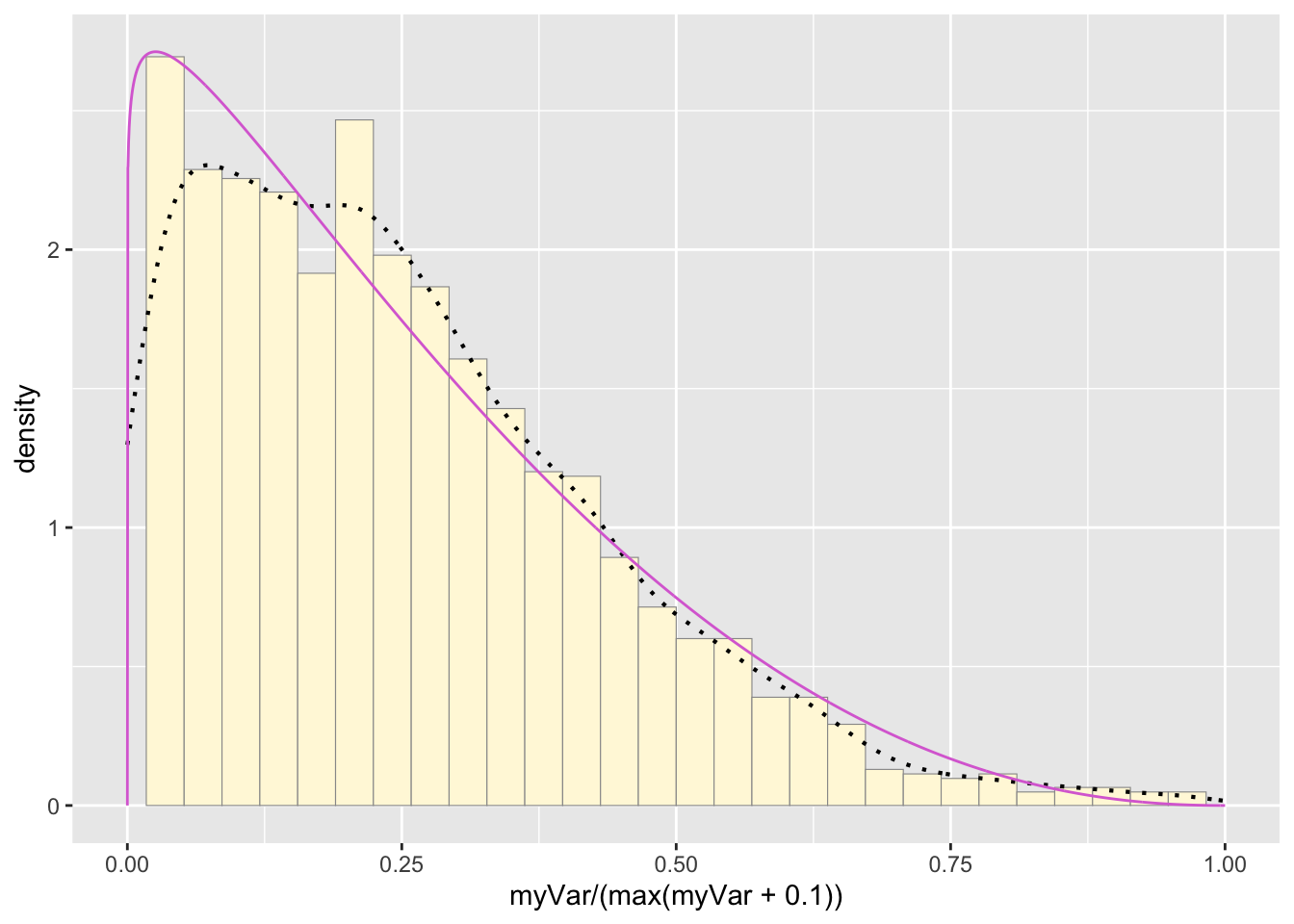
Question 2
Try it with your own data (starwars). Once the code is in and runs, try running this analysis on your own data.
#prepare our data
library(tidyverse)
z0 <- data.frame(starwars)
z <- na.omit(z0)
#view(z)
str(z)## 'data.frame': 29 obs. of 14 variables:
## $ name : chr "Luke Skywalker" "Darth Vader" "Leia Organa" "Owen Lars" ...
## $ height : int 172 202 150 178 165 183 182 188 228 180 ...
## $ mass : num 77 136 49 120 75 84 77 84 112 80 ...
## $ hair_color: chr "blond" "none" "brown" "brown, grey" ...
## $ skin_color: chr "fair" "white" "light" "light" ...
## $ eye_color : chr "blue" "yellow" "brown" "blue" ...
## $ birth_year: num 19 41.9 19 52 47 24 57 41.9 200 29 ...
## $ sex : chr "male" "male" "female" "male" ...
## $ gender : chr "masculine" "masculine" "feminine" "masculine" ...
## $ homeworld : chr "Tatooine" "Tatooine" "Alderaan" "Tatooine" ...
## $ species : chr "Human" "Human" "Human" "Human" ...
## $ films :List of 29
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "The Phantom Menace" ...
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "The Force Awakens"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi"
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi" "The Phantom Menace" "Attack of the Clones" ...
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi" "Attack of the Clones"
## ..$ : chr "The Empire Strikes Back"
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi"
## ..$ : chr "The Empire Strikes Back"
## ..$ : chr "Return of the Jedi" "The Force Awakens"
## ..$ : chr "Return of the Jedi"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones"
## ..$ : chr "The Phantom Menace"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones"
## ..$ : chr "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones"
## $ vehicles :List of 29
## ..$ : chr "Snowspeeder" "Imperial Speeder Bike"
## ..$ : chr
## ..$ : chr "Imperial Speeder Bike"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Tribubble bongo"
## ..$ : chr "Zephyr-G swoop bike" "XJ-6 airspeeder"
## ..$ : chr "AT-ST"
## ..$ : chr
## ..$ : chr "Snowspeeder"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Sith speeder"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Flitknot speeder"
## ..$ : chr
## $ starships :List of 29
## ..$ : chr "X-wing" "Imperial shuttle"
## ..$ : chr "TIE Advanced x1"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "X-wing"
## ..$ : chr "Jedi starfighter" "Trade Federation cruiser" "Naboo star skiff" "Jedi Interceptor" ...
## ..$ : chr "Naboo fighter" "Trade Federation cruiser" "Jedi Interceptor"
## ..$ : chr "Millennium Falcon" "Imperial shuttle"
## ..$ : chr "Millennium Falcon" "Imperial shuttle"
## ..$ : chr "X-wing"
## ..$ : chr
## ..$ : chr "Slave 1"
## ..$ : chr
## ..$ : chr "Millennium Falcon"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Naboo fighter" "H-type Nubian yacht" "Naboo star skiff"
## ..$ : chr
## ..$ : chr "Scimitar"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Jedi starfighter"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## - attr(*, "na.action")= 'omit' Named int [1:58] 2 3 8 12 15 16 18 19 22 27 ...
## ..- attr(*, "names")= chr [1:58] "2" "3" "8" "12" ...summary(z)## name height mass hair_color
## Length:29 Min. : 88.0 Min. : 20.00 Length:29
## Class :character 1st Qu.:172.0 1st Qu.: 75.00 Class :character
## Mode :character Median :180.0 Median : 79.00 Mode :character
## Mean :178.7 Mean : 77.77
## 3rd Qu.:188.0 3rd Qu.: 83.00
## Max. :228.0 Max. :136.00
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## skin_color eye_color birth_year sex
## Length:29 Length:29 Min. : 8.00 Length:29
## Class :character Class :character 1st Qu.: 31.00 Class :character
## Mode :character Mode :character Median : 46.00 Mode :character
## Mean : 51.29
## 3rd Qu.: 57.00
## Max. :200.00
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## gender homeworld species
## Length:29 Length:29 Length:29
## Class :character Class :character Class :character
## Mode :character Mode :character Mode :character
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## films.Length films.Class films.Mode
## 5 -none- character
## 4 -none- character
## 5 -none- character
## 3 -none- character
## 3 -none- character
## 1 -none- character
## 6 -none- character
## 3 -none- character
## 5 -none- character
## 4 -none- character
## 3 -none- character
## 5 -none- character
## 3 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 3 -none- character
## 2 -none- character
## 1 -none- character
## 3 -none- character
## 3 -none- character
## 3 -none- character
## 3 -none- character
## 2 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## vehicles.Length vehicles.Class vehicles.Mode
## 2 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## starships.Length starships.Class starships.Mode
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 5 -none- character
## 3 -none- character
## 2 -none- character
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 3 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character#plot histogram
p1 <- ggplot(data=z, aes(x=height, y=..density..)) +
geom_histogram(color="grey60",fill="cornsilk",linewidth=0.2)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.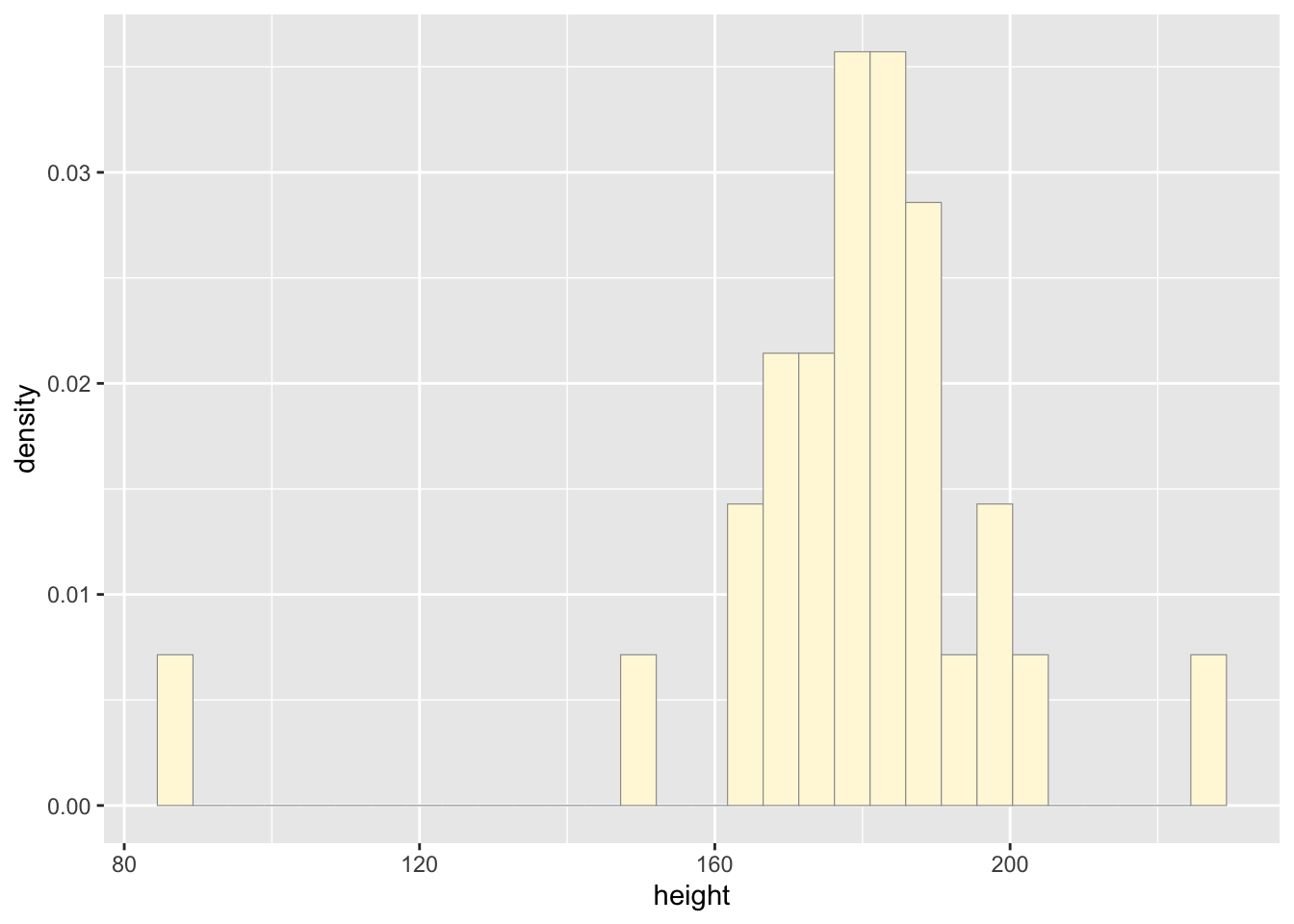
#add empirical density curve
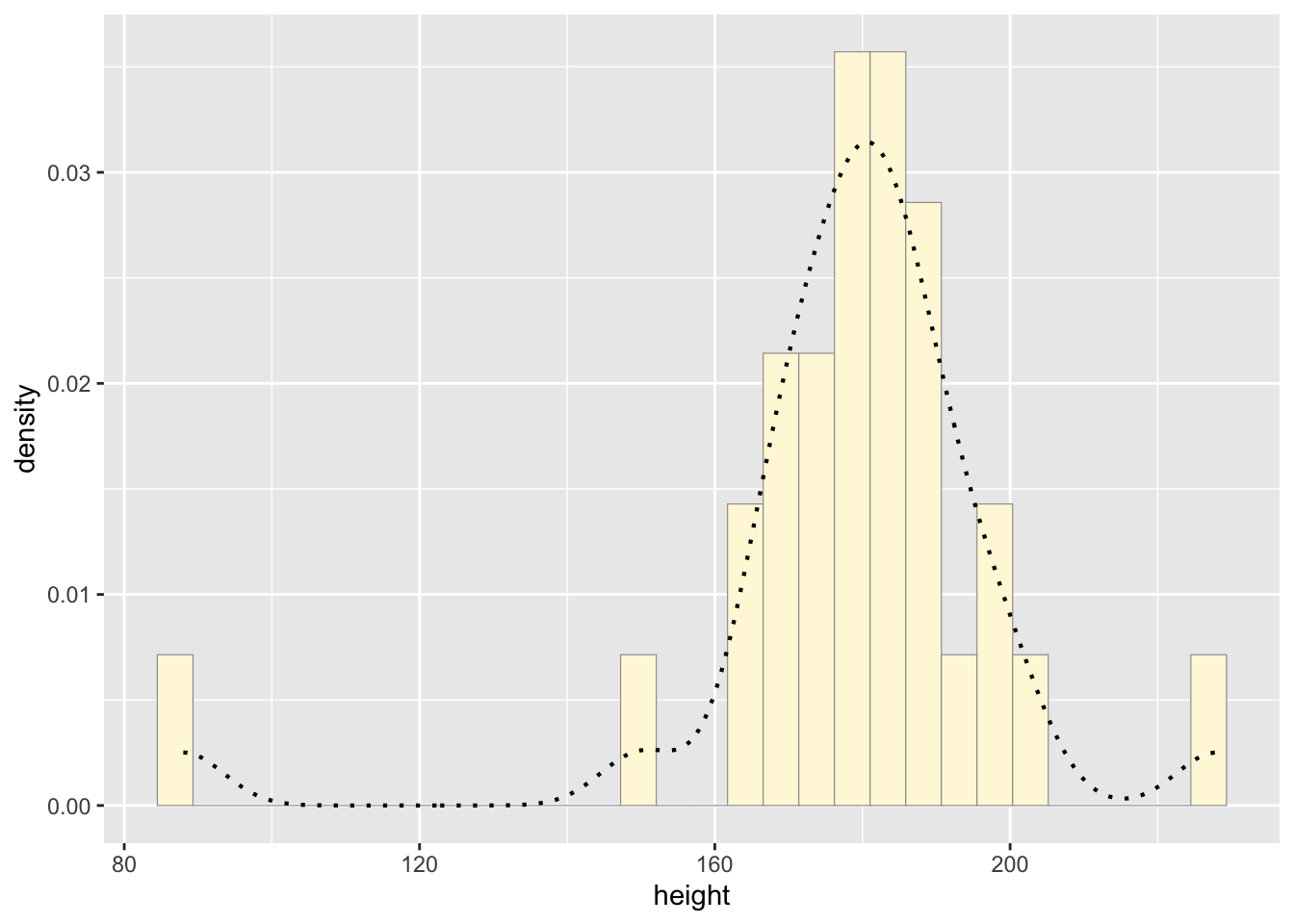
p1 <- p1 + geom_density(linetype="dotted",size=0.75)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.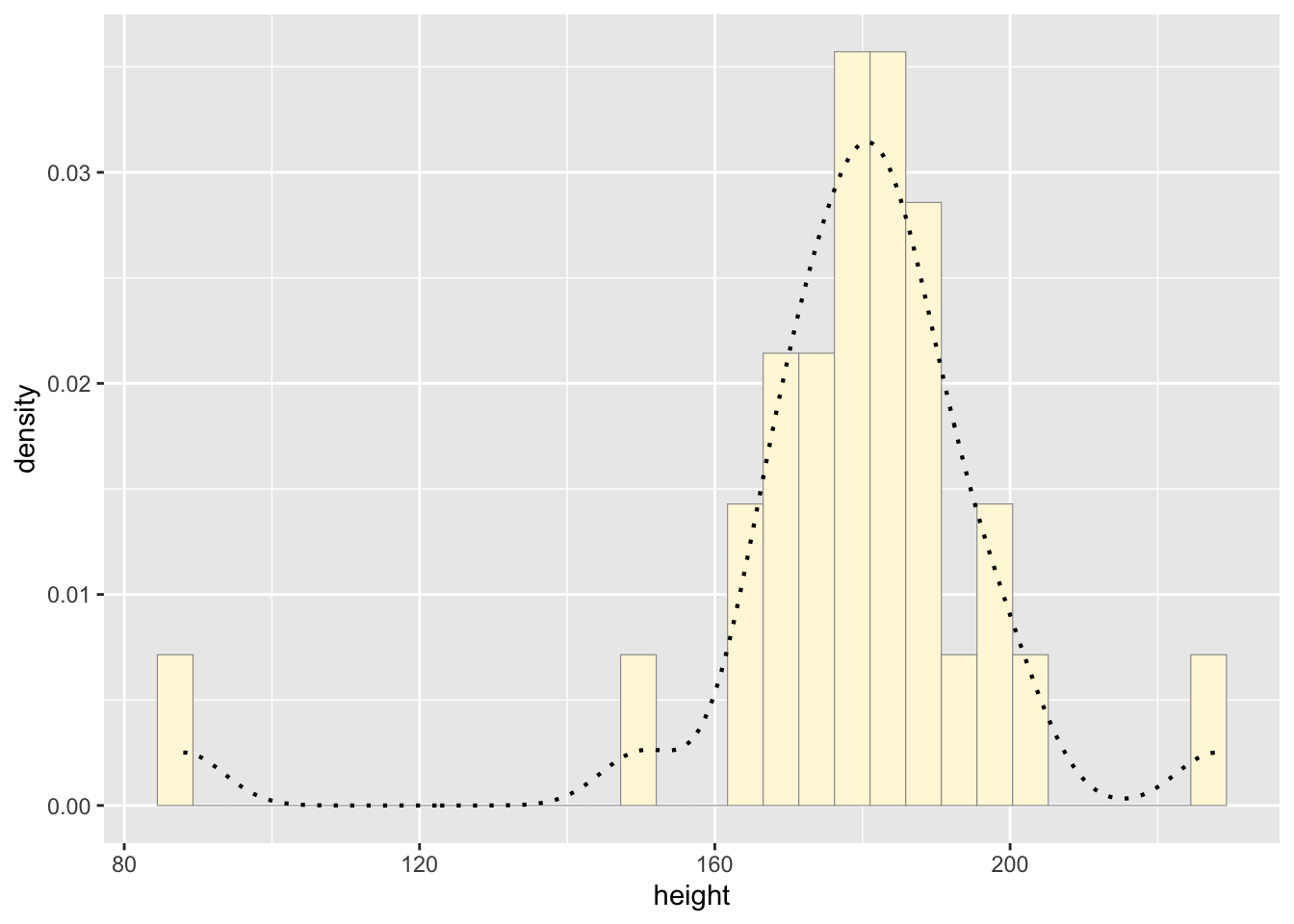
#get maximum likelihood parameters for normal
normPars <- fitdistr(z$height,"normal")
print(normPars)## mean sd
## 178.655172 22.009835
## ( 4.087124) ( 2.890033)str(normPars)## List of 5
## $ estimate: Named num [1:2] 179 22
## ..- attr(*, "names")= chr [1:2] "mean" "sd"
## $ sd : Named num [1:2] 4.09 2.89
## ..- attr(*, "names")= chr [1:2] "mean" "sd"
## $ vcov : num [1:2, 1:2] 16.7 0 0 8.35
## ..- attr(*, "dimnames")=List of 2
## .. ..$ : chr [1:2] "mean" "sd"
## .. ..$ : chr [1:2] "mean" "sd"
## $ n : int 29
## $ loglik : num -131
## - attr(*, "class")= chr "fitdistr"normPars$estimate["mean"] ## mean
## 178.6552#plot normal probability density
meanML <- normPars$estimate["mean"]
sdML <- normPars$estimate["sd"]
xval <- seq(0,max(z$height),len=length(z$height))
stat <- stat_function(aes(x = xval, y = ..y..), fun = dnorm, colour="red", n = length(z$height), args = list(mean = meanML, sd = sdML))
p1 + stat## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.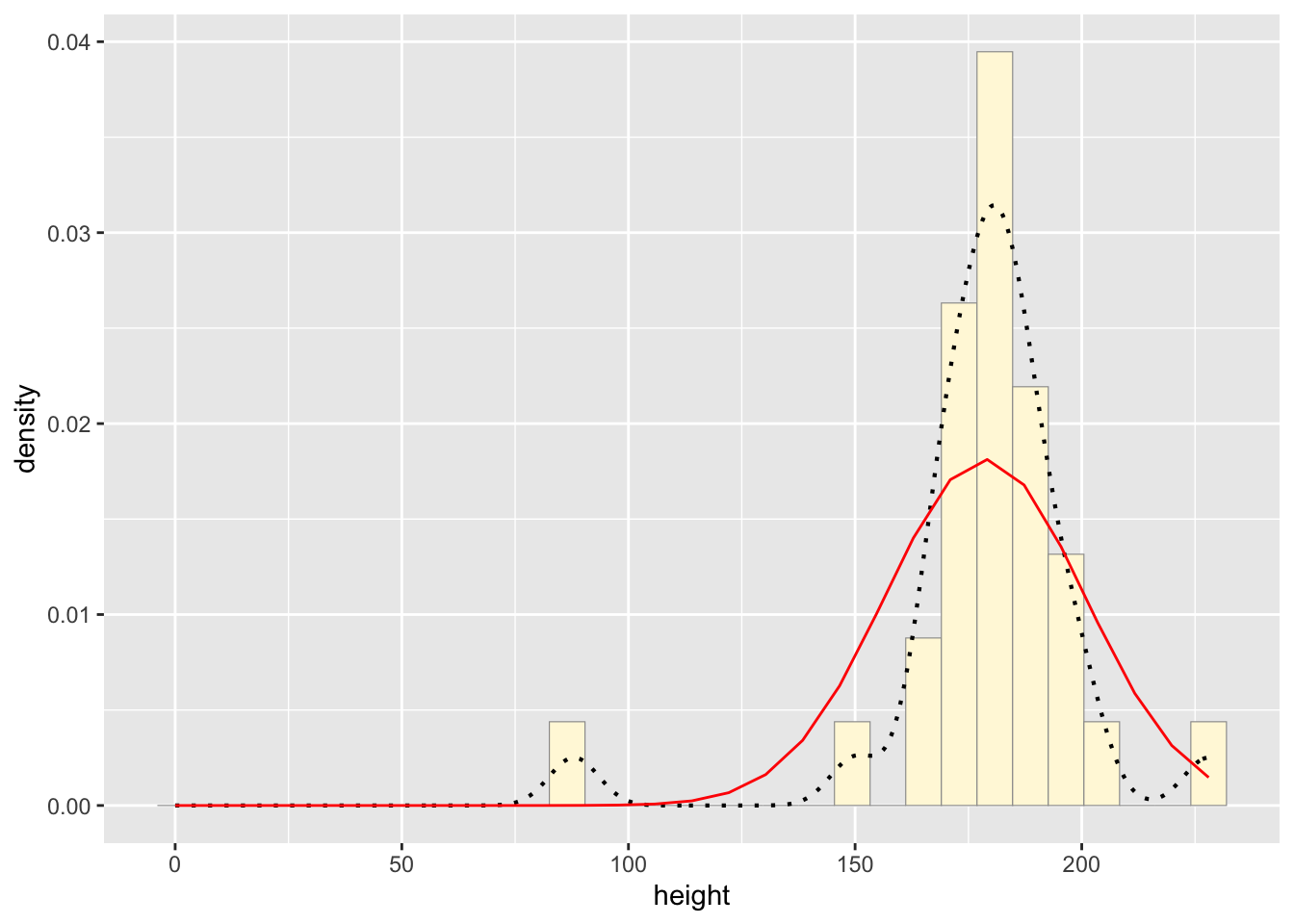
#plot exponential probability density
expoPars <- fitdistr(z$height,"exponential")
rateML <- expoPars$estimate["rate"]
stat2 <- stat_function(aes(x = xval, y = ..y..), fun = dexp, colour="blue", n = length(z$height), args = list(rate=rateML))
p1 + stat + stat2## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.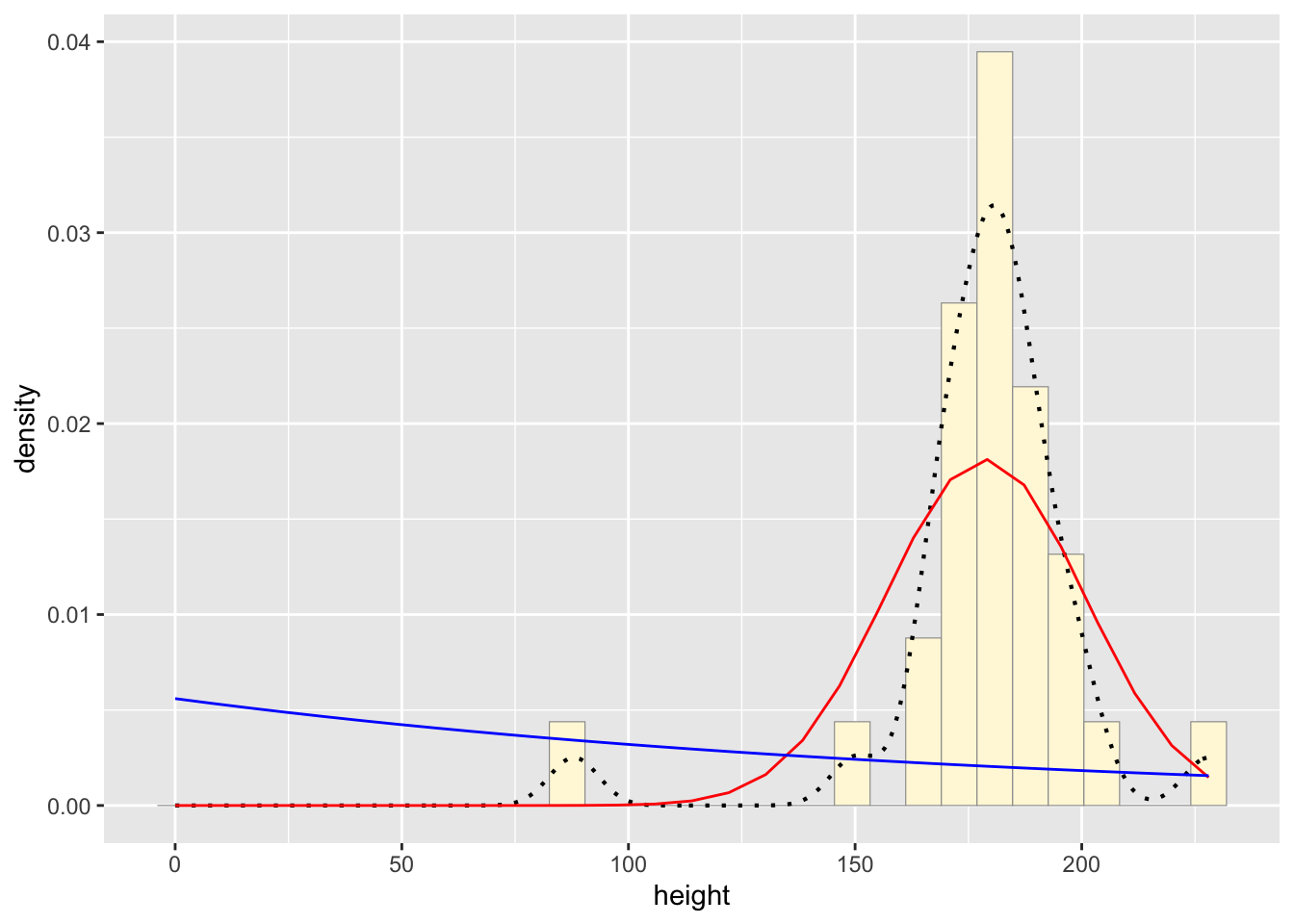
#plot uniform probability density
stat3 <- stat_function(aes(x = xval, y = ..y..), fun = dunif, colour="darkgreen", n = length(z$height), args = list(min=min(z$height), max=max(z$height)))
p1 + stat + stat2 + stat3## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.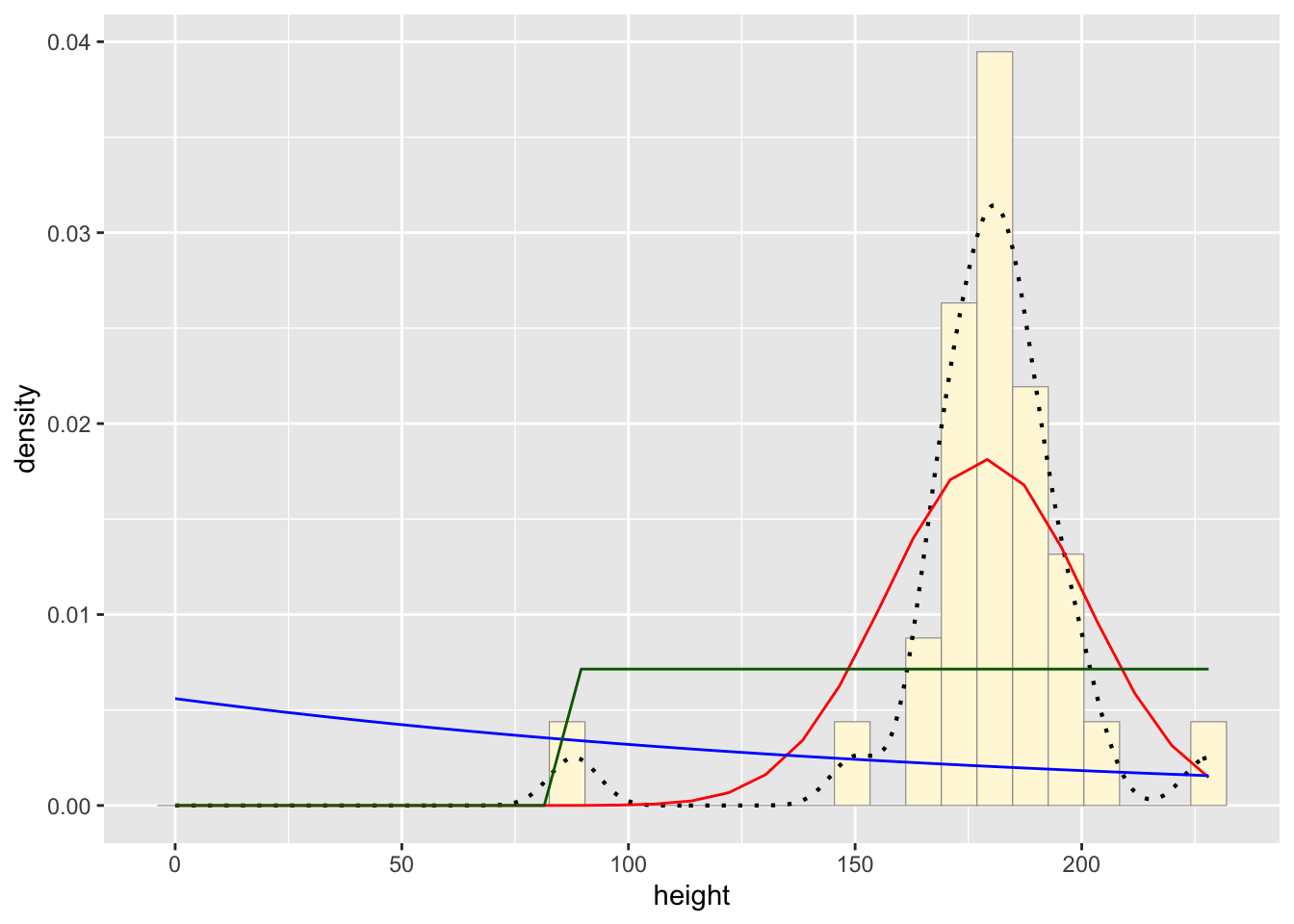
#plot gamma probability density
gammaPars <- fitdistr(z$height,"gamma")## Warning in densfun(x, parm[1], parm[2], ...): NaNs producedshapeML <- gammaPars$estimate["shape"]
rateML <- gammaPars$estimate["rate"]
stat4 <- stat_function(aes(x = xval, y = ..y..), fun = dgamma, colour="brown", n = length(z$height), args = list(shape=shapeML, rate=rateML))
p1 + stat + stat2 + stat3 + stat4## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.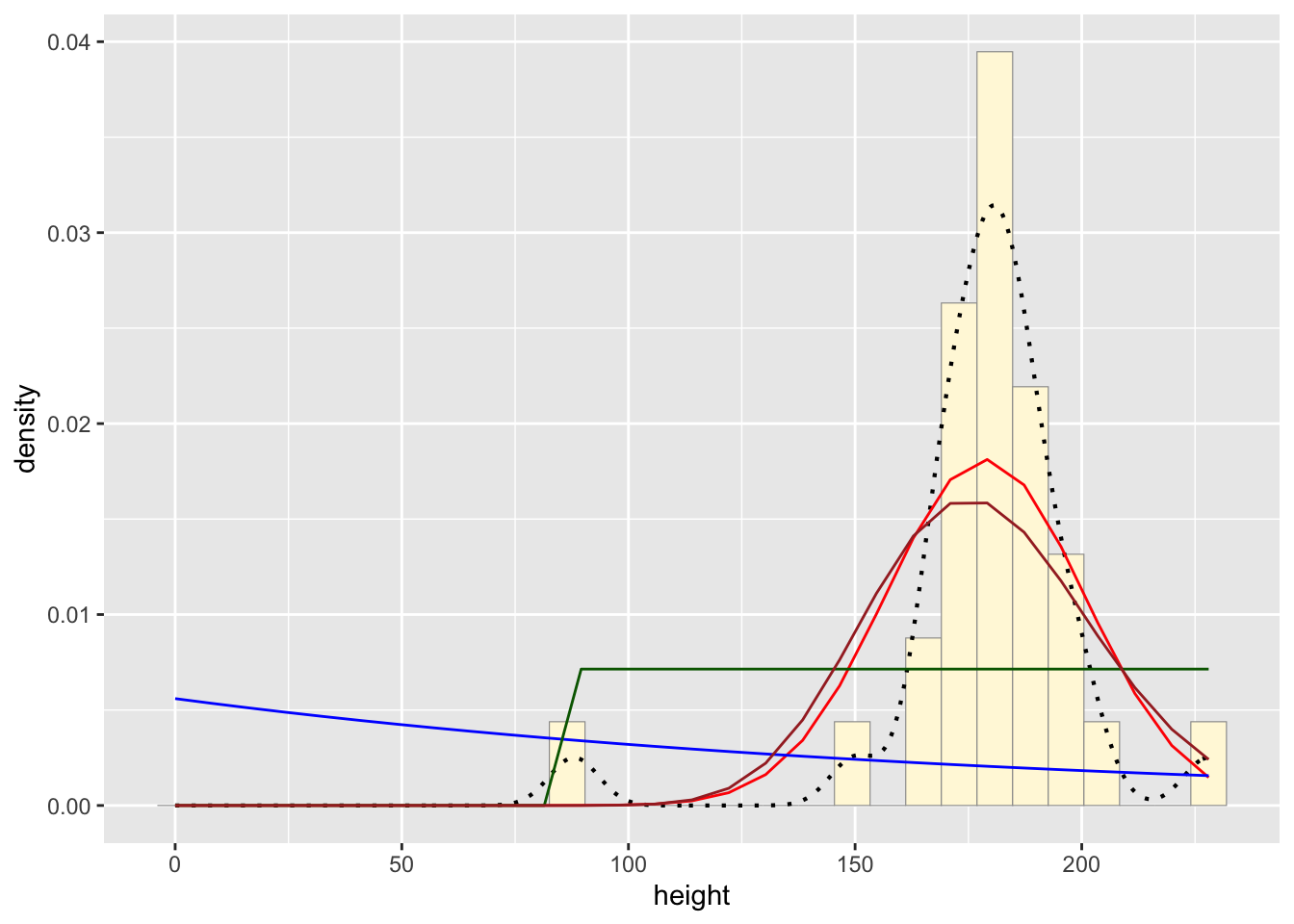
#plot beta probability density
pSpecial <- ggplot(data=z, aes(x=height/(max(height + 0.1)))) +
geom_histogram(color="grey60",fill="cornsilk",size=0.2) +
xlim(c(0,1)) +
geom_density(size=0.75,linetype="dotted")
betaPars <- fitdistr(x=z$height/max(z$height + 0.1),start=list(shape1=1,shape2=2),"beta")## Warning in densfun(x, parm[1], parm[2], ...): NaNs produced
## Warning in densfun(x, parm[1], parm[2], ...): NaNs producedshape1ML <- betaPars$estimate["shape1"]
shape2ML <- betaPars$estimate["shape2"]
statSpecial <- stat_function(aes(x = xval, y = ..y..), fun = dbeta, colour="orchid", n = length(z$height), args = list(shape1=shape1ML,shape2=shape2ML))
pSpecial + statSpecial## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.## Warning: Removed 2 rows containing missing values or values outside the
## scale range (`geom_bar()`).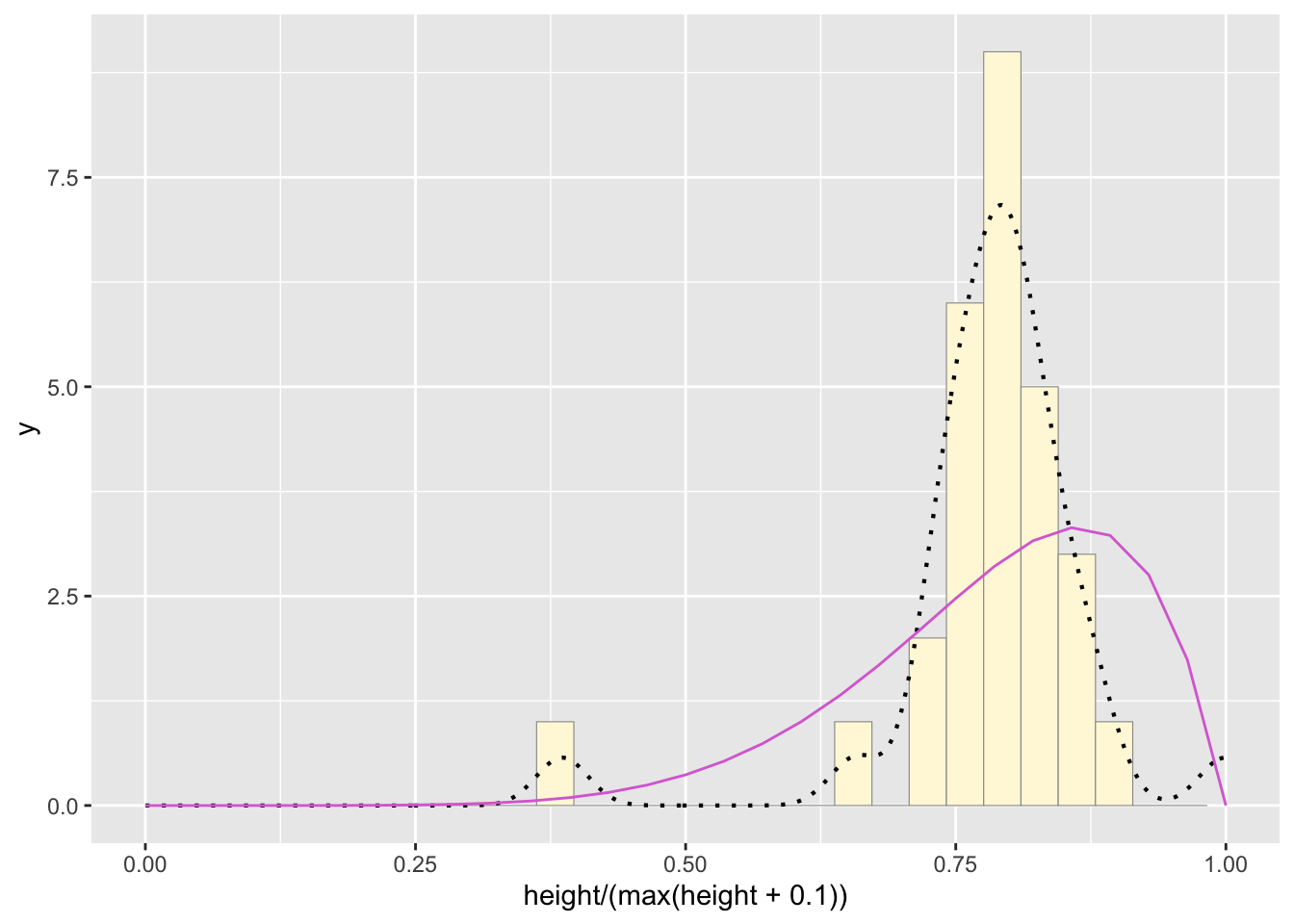
Question 3
Identify which distribution curve best fits the data.
Normal Probability Density curve best fits the data without
including bias like the beta density curve.
Question 4
Simulate a new data set.
No, the model is not
doing a good job because we used “rnorm” to generate a new dataset that
does not match the same distribution of data from the original.
Additionally, the sample size is small and might not be able to show the
best density distribution curve for the original data to be used for the
simulated data.
###Simulated Data
library(ggplot2)
library(MASS)
d <- rnorm(n=29,mean=178.66)
d <- data.frame(1:29,d)
names(d) <- list("ID","height")
d <- d[d$height>0,]
str(d)## 'data.frame': 29 obs. of 2 variables:
## $ ID : int 1 2 3 4 5 6 7 8 9 10 ...
## $ height: num 177 177 178 179 179 ...summary(d$height)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 176.5 177.8 178.8 178.6 179.4 180.6#plot histogram
p1 <- ggplot(data=d, aes(x=height, y=..density..)) +
geom_histogram(color="grey60",fill="cornsilk",linewidth=0.2)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.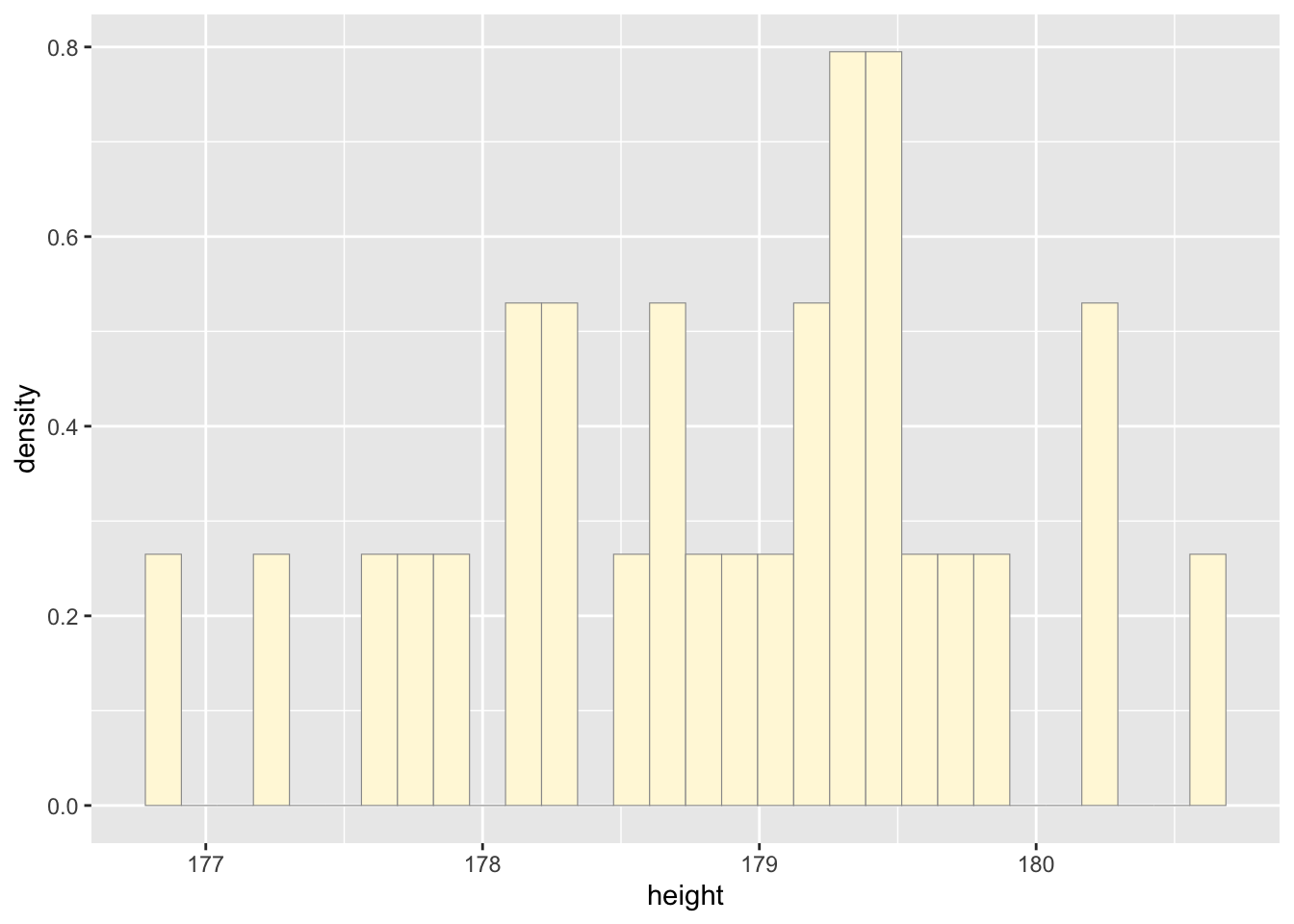
#add empirical density curve
p1 <- p1 + geom_density(linetype="dotted",size=0.75)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.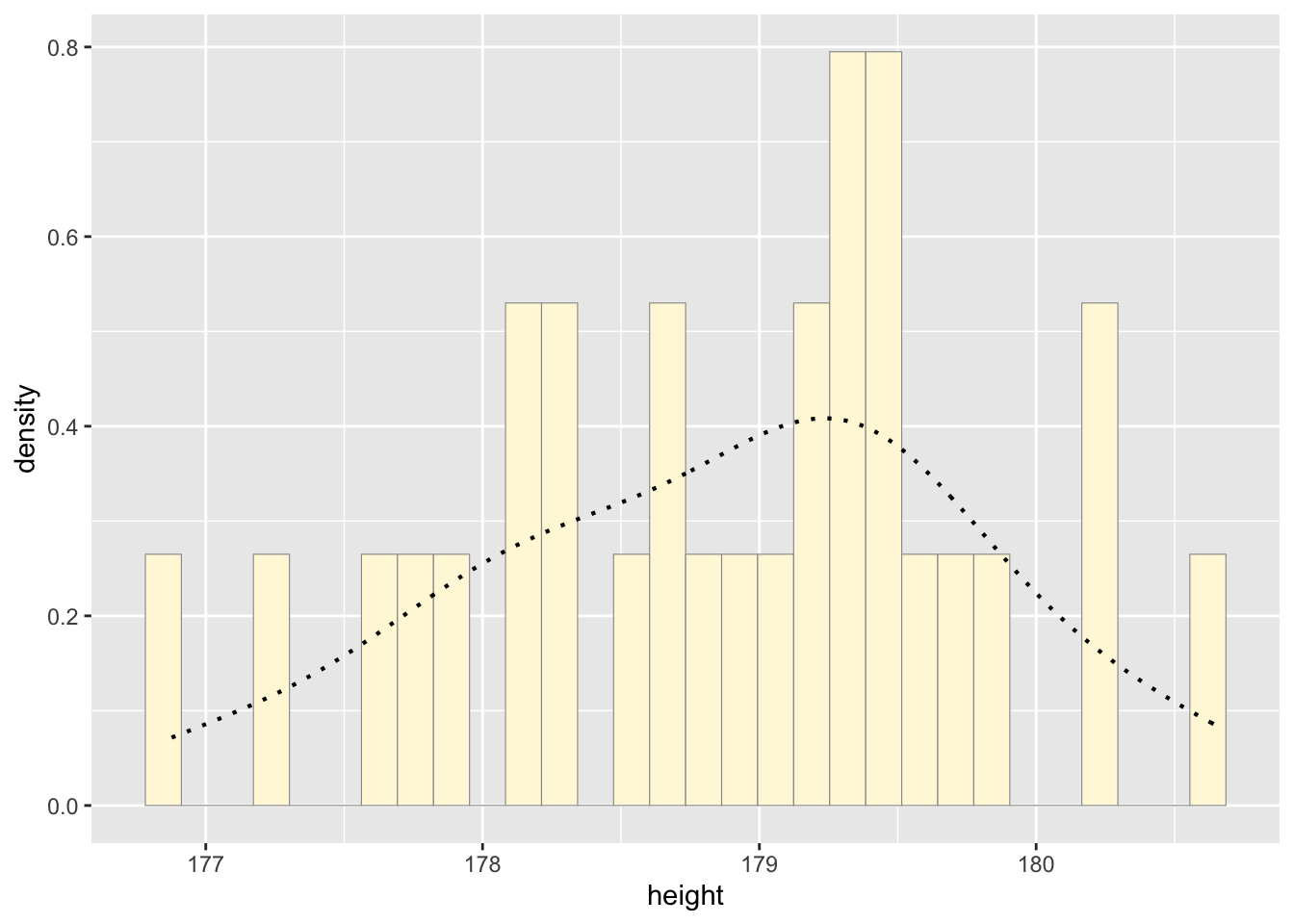
###Original Data
library(tidyverse)
z0 <- data.frame(starwars)
z <- na.omit(z0)
#view(z)
str(z)## 'data.frame': 29 obs. of 14 variables:
## $ name : chr "Luke Skywalker" "Darth Vader" "Leia Organa" "Owen Lars" ...
## $ height : int 172 202 150 178 165 183 182 188 228 180 ...
## $ mass : num 77 136 49 120 75 84 77 84 112 80 ...
## $ hair_color: chr "blond" "none" "brown" "brown, grey" ...
## $ skin_color: chr "fair" "white" "light" "light" ...
## $ eye_color : chr "blue" "yellow" "brown" "blue" ...
## $ birth_year: num 19 41.9 19 52 47 24 57 41.9 200 29 ...
## $ sex : chr "male" "male" "female" "male" ...
## $ gender : chr "masculine" "masculine" "feminine" "masculine" ...
## $ homeworld : chr "Tatooine" "Tatooine" "Alderaan" "Tatooine" ...
## $ species : chr "Human" "Human" "Human" "Human" ...
## $ films :List of 29
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "The Phantom Menace" ...
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "Revenge of the Sith" ...
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi" "The Force Awakens"
## ..$ : chr "A New Hope" "The Empire Strikes Back" "Return of the Jedi"
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi" "The Phantom Menace" "Attack of the Clones" ...
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi" "Attack of the Clones"
## ..$ : chr "The Empire Strikes Back"
## ..$ : chr "The Empire Strikes Back" "Return of the Jedi"
## ..$ : chr "The Empire Strikes Back"
## ..$ : chr "Return of the Jedi" "The Force Awakens"
## ..$ : chr "Return of the Jedi"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones"
## ..$ : chr "The Phantom Menace"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "The Phantom Menace" "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones"
## ..$ : chr "Attack of the Clones" "Revenge of the Sith"
## ..$ : chr "Attack of the Clones"
## $ vehicles :List of 29
## ..$ : chr "Snowspeeder" "Imperial Speeder Bike"
## ..$ : chr
## ..$ : chr "Imperial Speeder Bike"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Tribubble bongo"
## ..$ : chr "Zephyr-G swoop bike" "XJ-6 airspeeder"
## ..$ : chr "AT-ST"
## ..$ : chr
## ..$ : chr "Snowspeeder"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Sith speeder"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Flitknot speeder"
## ..$ : chr
## $ starships :List of 29
## ..$ : chr "X-wing" "Imperial shuttle"
## ..$ : chr "TIE Advanced x1"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "X-wing"
## ..$ : chr "Jedi starfighter" "Trade Federation cruiser" "Naboo star skiff" "Jedi Interceptor" ...
## ..$ : chr "Naboo fighter" "Trade Federation cruiser" "Jedi Interceptor"
## ..$ : chr "Millennium Falcon" "Imperial shuttle"
## ..$ : chr "Millennium Falcon" "Imperial shuttle"
## ..$ : chr "X-wing"
## ..$ : chr
## ..$ : chr "Slave 1"
## ..$ : chr
## ..$ : chr "Millennium Falcon"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Naboo fighter" "H-type Nubian yacht" "Naboo star skiff"
## ..$ : chr
## ..$ : chr "Scimitar"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr "Jedi starfighter"
## ..$ : chr
## ..$ : chr
## ..$ : chr
## ..$ : chr
## - attr(*, "na.action")= 'omit' Named int [1:58] 2 3 8 12 15 16 18 19 22 27 ...
## ..- attr(*, "names")= chr [1:58] "2" "3" "8" "12" ...summary(z)## name height mass hair_color
## Length:29 Min. : 88.0 Min. : 20.00 Length:29
## Class :character 1st Qu.:172.0 1st Qu.: 75.00 Class :character
## Mode :character Median :180.0 Median : 79.00 Mode :character
## Mean :178.7 Mean : 77.77
## 3rd Qu.:188.0 3rd Qu.: 83.00
## Max. :228.0 Max. :136.00
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## skin_color eye_color birth_year sex
## Length:29 Length:29 Min. : 8.00 Length:29
## Class :character Class :character 1st Qu.: 31.00 Class :character
## Mode :character Mode :character Median : 46.00 Mode :character
## Mean : 51.29
## 3rd Qu.: 57.00
## Max. :200.00
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## gender homeworld species
## Length:29 Length:29 Length:29
## Class :character Class :character Class :character
## Mode :character Mode :character Mode :character
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
##
## films.Length films.Class films.Mode
## 5 -none- character
## 4 -none- character
## 5 -none- character
## 3 -none- character
## 3 -none- character
## 1 -none- character
## 6 -none- character
## 3 -none- character
## 5 -none- character
## 4 -none- character
## 3 -none- character
## 5 -none- character
## 3 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 3 -none- character
## 2 -none- character
## 1 -none- character
## 3 -none- character
## 3 -none- character
## 3 -none- character
## 3 -none- character
## 2 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## vehicles.Length vehicles.Class vehicles.Mode
## 2 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## starships.Length starships.Class starships.Mode
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 5 -none- character
## 3 -none- character
## 2 -none- character
## 2 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 3 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 1 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character
## 0 -none- character#plot histogram
p1 <- ggplot(data=z, aes(x=height, y=..density..)) +
geom_histogram(color="grey60",fill="cornsilk",linewidth=0.2)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.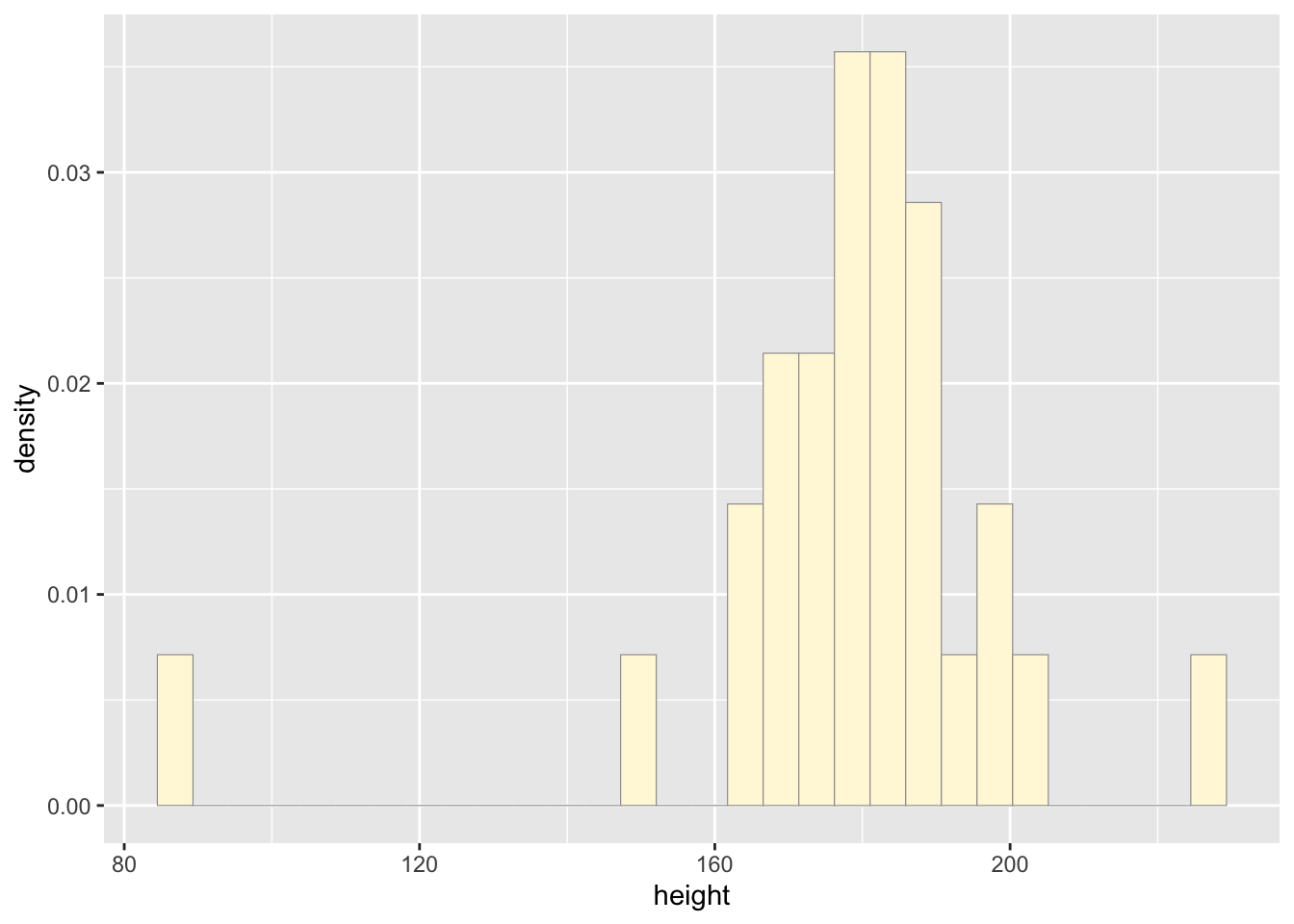
#add empirical density curve
p1 <- p1 + geom_density(linetype="dotted",size=0.75)
print(p1)## `stat_bin()` using `bins = 30`. Pick better value with
## `binwidth`.